A Day with Microbial Taxanomist
Hello Steemians,
It’s going to be the first article, so I thought why not to share something what generally I do or a Microbial Taxonomist generally do in lab. Taking you on a journey with Microbial Taxanomist. A Taxanomist basically groups the organism based on their features and characteristics and a Microbial Taxanomist generally works with the microbes.
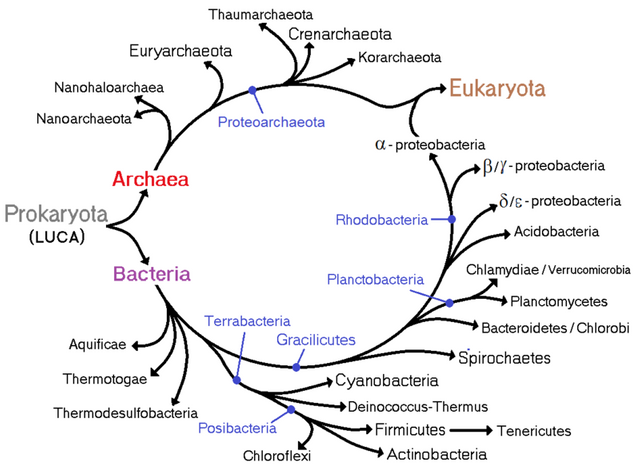
From the beginning in 1676 when Antonie van Leeuwenhoek observed the first bacteria through the single lens microscope several other reports came across for the existence of bacterial species. Ernst Haeckel, Carl woese, R H Whittaker and many more contributed to build the phylogenetic tree of life with all the kingdoms in it. After it was clear that Bacteria, Archaea and Eukaryotes are in different line of classification, Bacteria was sub-divided into two groups: According to their cell wall composition which can be differentiated based on Gram staining. Considering the gram staining sub-group division Bergey’s divided them in four different divisions: Gracilicutes, Firmacutes, Mollicutes and Mendocutes.
- Gracilicutes (Photosynthetic or non-photosynthetic gram-negative)
- Firmacutes (Gram positive including the Actinobacter)
- Mollicutes (Gram variable, cell wall composition vary through generations eg; Mycoplasma)
- Mendocutes (Archaebacteria group)
This is how microbe are being divided into their respective genus and species and the Taxanomy starts.
To start with one must have a microbial source or a niche from where you want to find a bacterial species. As you know bacteria are present everywhere so why does a specific site matters for it finding. The answer is pretty clear, all those bacteria which are present in the surrounding are well studied and to find a new one you must look for a place where no one explored yet. So, the source site matters a lot.
The whole journey starts with finding a potential source for microbes to get a new one and to add it to the literature. The sampling could be of soil, water, waste, human gut etc. it could be anything. The first thing to keep in mind which group you are looking for and then accordingly go for its niche and if you are just going for a novel one better to look in a place no one ever reached. The sterility of the whole system should be taken care off, because its microbe you can catch them anywhere. The samples are taken in a sterilized containers and some time we also try to take the natural environment in which the microbes are surviving. Because it is something very important as the microbes you want to find may not survive the condition and nutrition you provide.
“Almost 90% of the microbial population could not survive after being taken out of their actual environment”
So, sometime it is very necessary to try mimic the whole condition or take the natural environment and try to subculture bacteria in that only. But this also leads to the forfeit of microbial species. Several advancement in science can let you grow the bacteria in the same niche they live in. NovoBiotic Pharmaceutical has developed an Isolation chip called i-chip which can be inserted into the ground and its several wells can have different bacteria species distributed and growing in it. The bacterial samples are now plated onto the media plates for their growth. The media is the food source or the nutrition for microbes and these media supplement varies accordingly. As for the main source sugar and carbohydrate is necessary and apart from it sometime several vitamins and proteins are also required and all these are based on the area from where you are trying to find bacteria. Let’s take an example; if you want to isolate a bacteria from human gut or dental carries you must add blood component in your media to let the bacteria grow outside also. The media will provide the exact nutritional requirement to let it grow. Apart from the nutrient content the pH also matters as there are specific group of bacteria which grows only at a specific pH and cannot survive the change in pH.
With media there are some other things also which are the main components for the survival of these microbial species (The environment). Bacteria are grouped according to the oxygen tolerance and in tolerance. Some of them can survive in oxygen called Aerobic, some cannot survive the presence of oxygen called Obligate Anaerobic and some can withstand the presence of oxygen and can grow with or without the presence of oxygen are called Facultative Anaerobes.
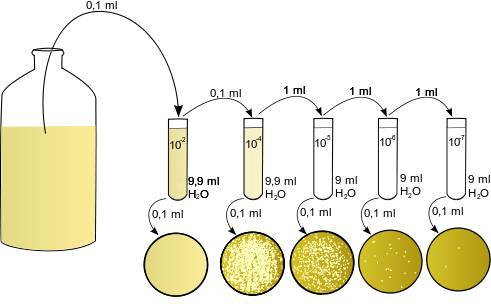
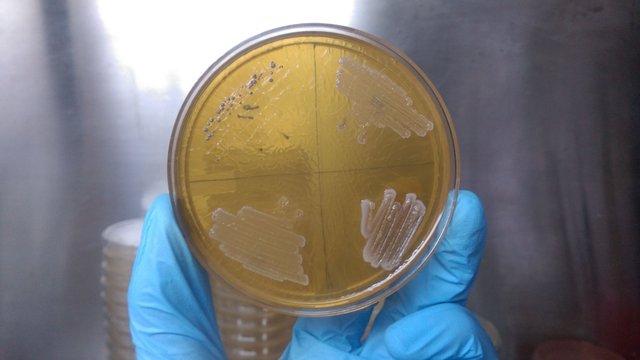
So, after the sampling is done the sample is been processed for the inoculation. A single gram of soil or 1ml of a sewage water can contain several hundreds or thousands of bacteria so to culture them dilution of the sample is necessary and it’s been done by serial dilution where 1/10th part goes to the next tube and diluting the whole sample in a gap of 10 folds. After the dilution is done the sample were quickly spreaded on to the nutrient plates. As we discussed for the different genera we have to prepare different media so the sample were plated onto the different media plates and we can end up with getting a wide variety of microorganisms.
After the plating or inoculation of the samples the agar plates and media flasks or bottles are kept in the incubators where the temperature is maintained to grow the microbes. Sometime we have to try a different range of temperature just to see which bacteria can survive up to an extreme temperature and different temperature incubators also helps to maintain the required temperature. In most of the cases usually bacteria have a doubling time or generation time of 20-40 minutes but can also vary. Depending on the bacterial species they can have the prolonged generation time or some time proper nutrition and environment also plays a crucial role in the growth of bacterial species. These culture plate should be monitored regularly up to 3-4 days and any new or unique bacterial colony can be picked for the further study. The bacterial growth is exponential they divide like 2-4-8-16-32-64-128-256-512-1024 and so on. So, within nine generation of 20 minutes the cell number increases from 1 to 1000. After an overnight incubation you can find lots of bacterial culture on your plate, so better to make a proper dilution. That will help in the separate and single bacterial colonies.
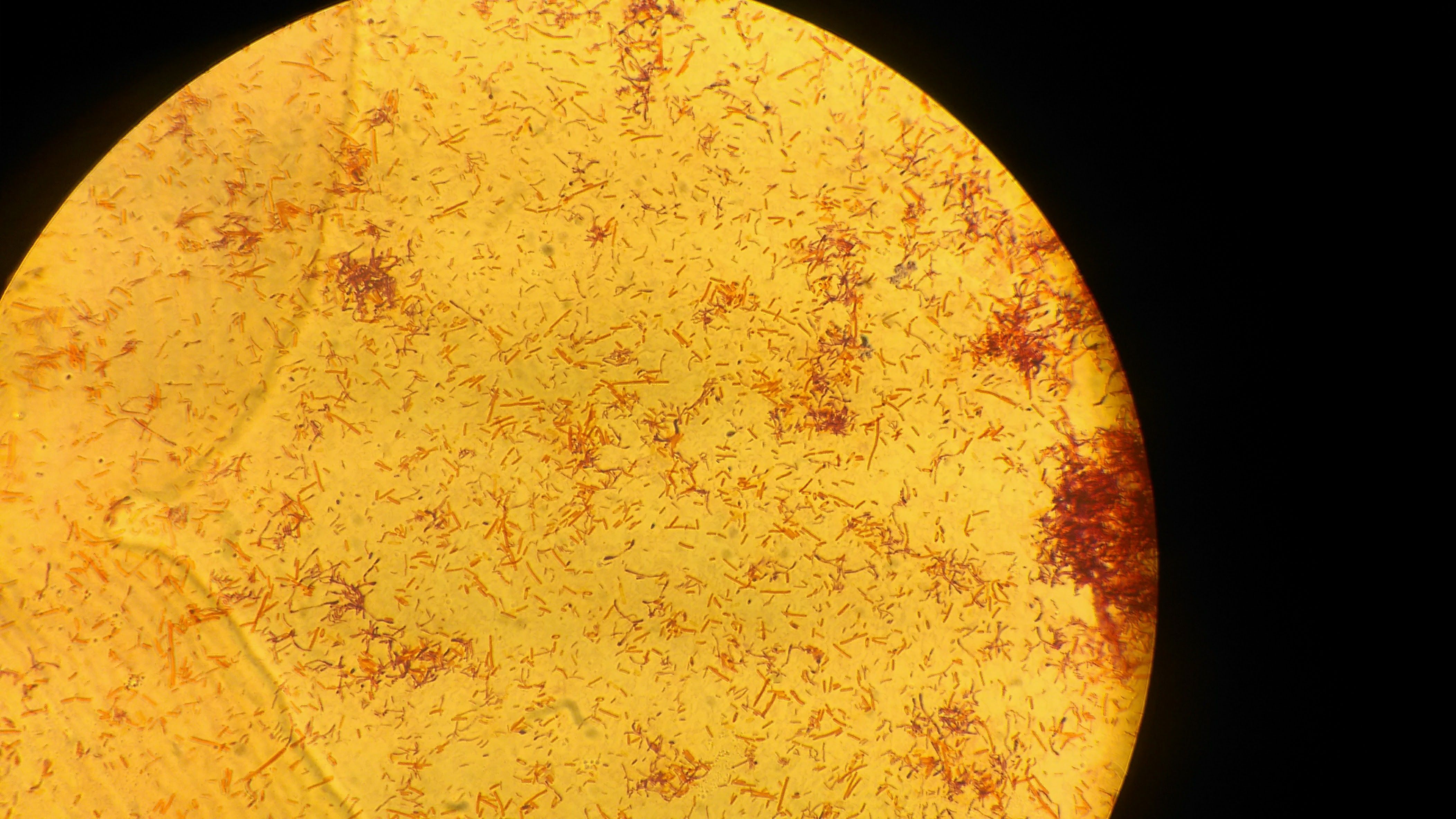
Gram negative rods observed under 100X immersion
Now comes the characterization part of the bacteria which are being cultured. As I earlier described, they are first characterized based on the gram staining. Working in a microbiology lab will teach you some of the characteristics which are helpful in distinguishing between the gram positive and gram negative bacteria by just looking the bacterial colonies without performing the gram stain. I would not recommend this but it helps many time in excluding the unwanted bacterial population. The gram positive bacteria will be having king rough surface and whereas the gram negative bacteria will be having quite smooth and shiny surface and the basic fundamental behind this is the gram negatives are having higher amount of lipopolysaccharide in their cell wall and make them shiny and smooth. But it is not always true, if you are confused better to do the staining and find out. Gram staining named after the Danish Bacteriologist Hans Christian Gram who gave the technique to stain the cell wall and differentiate between bacteria.
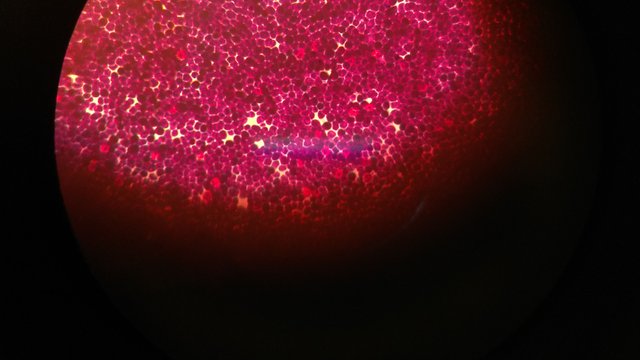
Gram positive cocci observed under 100X oil immersion.
Peptidoglycans present in the bacterial membrane get stained by the complex of crystal violet and iodine. As the gram positive bacteria have a thick layer it captures the dye strongly and shows violet color while the gram negative bacterial contain very thin layer of peptidoglycan which on further treatment with the alcohol get dissolved and hence cannot retain the violet color and which was further stained through the safranin, which gives the pink color to the bacterial cells.
Next Generation Sequencing (NGS)
The important and most widely used technique to identify the novel bacteria is genome sequencing. All species has a conserved sequence also called as house-keeping genes which are present throughout generations and are best to identify the new species. In bacteria the conserved sequence is 16S ribosomal gene which is merely 1500 base pairs of nucleotide (ATCG) and has about seven different universal primers for the sequencing of these genes. There is also an option of whole genome sequencing but that is bit time taking and really costly only be done when it’s necessary. Sequencing a 16SrRNA gene sequence requires two primers which can anneal through the whole stretch and give the sequence. It’s basically done by Polymerase chain reaction (PCR) some of you may have heard of it or may be in my coming articles I will explain it. What PCR does is it amplifies the gene copy and result in the huge amount of desired gene sequence.
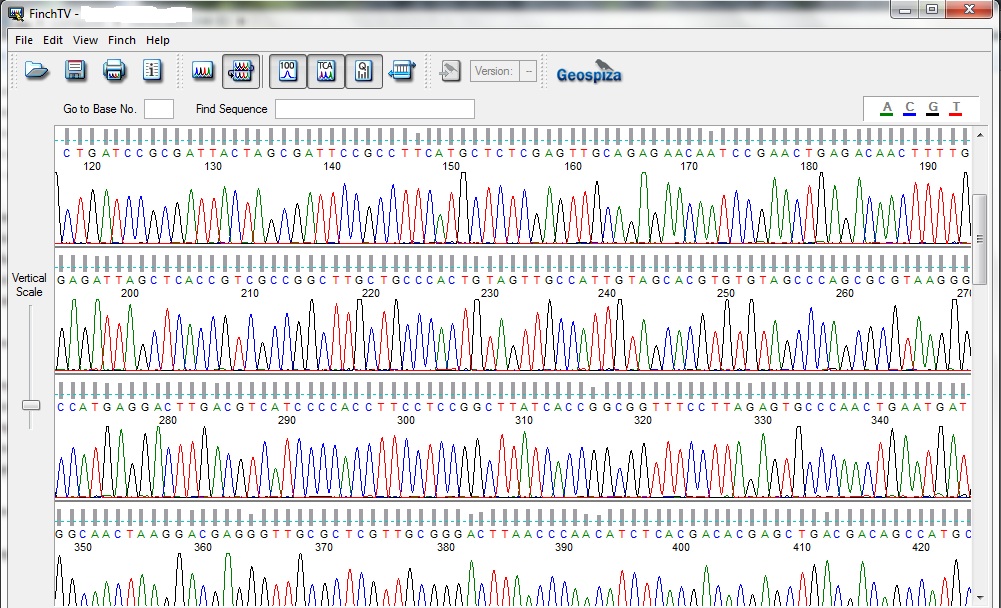
Chromatogram of Sanger Sequencing
Through gel electrophoresis and further purification the amplified gene can be procured which further subjected to sequencing which shows the nucleotide chromatograms. The chromatograms suggests the sequential addition of nucleotide onto the single strand the principle of this sequencing is based on Sanger sequencing. It’s a step wise addition where addition of any nucleotide will give a respective colored fluorescence and show in the display as a chromatogram peak. The collected gene sequence then aligned with the database called Basic Local Alignment Search Tool (BLAST). As the gene is conserved about 90% of the gene always carries the same nucleotide stretch but the percentage similarity which differentiates the bacteria from the other known bacteria is less than 97% similarity. 95-97% similarity can be acclaimed as new species of that genus but 93-94% can be given to a new genus, it’s all on the differentiability of the isolated bacteria.
With the BLAST file or aligned file you can build a phylogenetic tree using several applications like MEGA, EzEditer, PHYLIP etc. The phylogenetic tree or can be called as evolutionary tree suggests the emergence of the new species and to which genus the new species is similar in characters and further named under that family. The phylogenetic tree have several branches and each branch descends from a nod shows the new species. This usually suggests whether you have found a novel species of bacteria or not and later on there are so many other biochemical tests certain protein profiling and all to verify the new species. There is something which I want to share is to differentiate between two very similar species or sub-species of same genus DNA-DNA hybridization can also be done. It usually deals with the hybridization of the whole genomic DNA of one species onto the other Genomic DNA which is radioactive labelled and on hybridization give a signal in percentage similarity. This I will definitely share with you people, the whole experiment how does it worked.
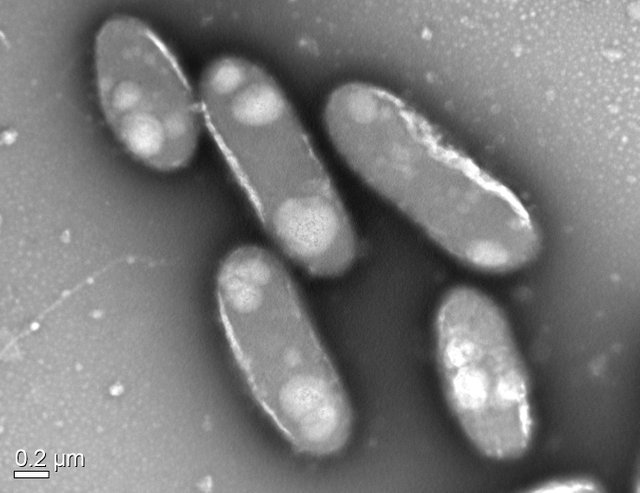
Transmission Electron Micrograph Image of a Bacterial species
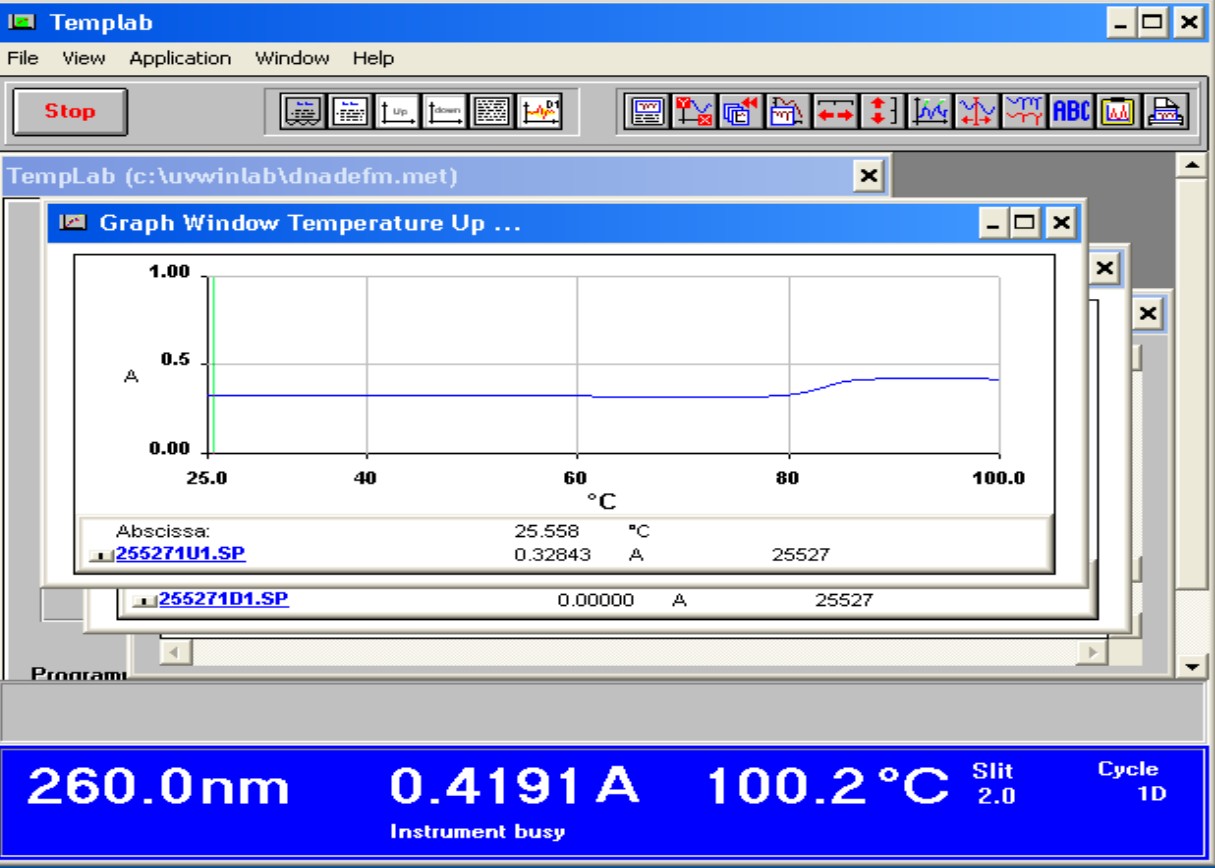
G+C content Profile
There is much more a taxonomist do, but certainly not in a single day. I’ll be sharing them all just stay fit and be aware of the harmful microbes. They can be deadly too.
Images which are not sourced are taken by me and do not violates any copyright.
Hope you like the read, please upvote and follow my page for reading more of such articles on microbiology.
Thank You
Welcome to Steemit @micro24! Great to see some more microbiology articles on here. Looking forward to your next posts.
Thankyou @tking77798
Will try to produce good stuff here.
You misspelled your occupation 4 times in the opening paragraph and 1 in the title 😃
Yeah, its Taxonomist sorry for that and thanks for correcting it.
Welcome to Steemit @micro24! I am interested in similar topics. Much of my work was cloning and sequencing 16S rRNA of bacteria in caves and producing phylogenetic trees. I'll follow you and perhaps we will find each other's posts interesting.
Thank You @qiyi
This is what I was told to and was expecting, to find people of same field from different places of world. Following you back and love to see your articles.
Hi.. Happy to see someone from India writing quality science articles :) Are you on discord?
Thankyou @dexterdev
Just made the account on discord and will be joining steemstem group.
great write up, loved learning about the different techniques for identification !
Thankyou @mcfarhat
Will be explaining them in detail in future as some of the readers are not aware of all the techniques.
Very well written article. How difficult is it to discover new species of bacteria? Is it still a common event to discover a new species?
I see. Not so difficult to find a different signal, but difficult to accurately identify exactly what is making that signal. I understand even if I didn’t manage to restate it very elegantly!
@flyyingkiwi you can see my latest post, its about the 16S rRNA gene sequencing and identifying a novel bacteria species.
Easy to find novel 16S rRNA. Difficult to isolate the bug responsible and create a type strain to make it an official species.
Thank you @flyyingkiwi and @effofex
16S rRNA gene sequencing is best to identify one but to find one from the source is like mining. You have to find the one among hundereds or even thousand but if you know how to find them and what characterstics a novel one could possess, it would be much easier to find the novel species.
Hi @micro24. You need to cite your article's references to get an upvote from the steemstem (there is no guarantee though). Your explanation was quite extensive and even though I am not familiar with some of the terminologies, it was fun. Kudos.
Thanks for the suggestion,
Sorry for that, will be putting some references from next time so to make you understand what you didn't get.
Is that the four divisions of microbes by Bergey's you didn't get or some thing else?
@micro24 : You can edit the post now too with references. If you want to :)
No problem. Yeah, one of the things would be that. I've never heard gracilicutes, firmacutes, mollicutes and mendocutes before but the most things I struggled to understand was the Next Generation Sequencing (NGS).
Next time surely will be adding the reference 😊
Congratulations @micro24! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on any badge to view your own Board of Honor on SteemitBoard.
For more information about SteemitBoard, click here
If you no longer want to receive notifications, reply to this comment with the word
STOP